2.4.3. Information Module
This page provides a summary of the functions available in the information module.
Currently, the following functions are available:
2.4.3.1. Entropy
One of the most common ways to represent information from a data source is through the measurement of entropy. In this sub-module we have included some implementations of measuring entropy from a signal (permutation entropy) and persistence diagram (persistent entropy). Entropy is calculated as
where p(x) is the probability of data type x, which is an element of the finite set of data types X. Additional, the log is base 2 so that the entropy as units of information. Persistent entropy is normalized on a scale from [0,1] with no units as
where # denotes the number of data types in X.
2.4.3.1.1. Permutation Entropy
Permutation entropy uses the data types of permutations (see Permutation Sequence Generation for details on permutations) and calculates the probability of each as their respective occurance in a time series.
- teaspoon.SP.information.entropy.PE(ts, n=5, tau=10, normalize=False)[source]
This function takes a time series and calculates Permutation Entropy (PE).
- Parameters:
ts (array) – Time series (1d).
n (int) – Permutation dimension. Default is 5.
tau (int) – Permutation delay. Default is 10.
- Kwargs:
normalize (bool): Normalizes the permutation entropy on scale from 0 to 1. defaut is False.
- Returns:
PE, the permutation entropy.
- Return type:
(float)
Example:
import numpy as np
t = np.linspace(0,100,2000)
ts = np.sin(t) #generate a simple time series
from teaspoon.SP.information.entropy import PE
h = PE(ts, n = 6, tau = 15, normalize = True)
print('Permutation entropy: ', h)
Output of example:
Permutation entropy: 0.4350397222113192
2.4.3.1.2. Multi-scale Permutation Entropy
Multi-scale Permutation Entropy applies the permutation entropy calculation over multiple time scales (delays).
- teaspoon.SP.information.entropy.MsPE(ts, n=5, delay_end=200, plotting=False, normalize=False)[source]
- This function takes a time series and calculates Multi-scale Permutation Entropy (MsPE)
over multiple time scales
- Parameters:
ts (array) – Time series (1d).
n (int) – Permutation dimension. Default is 5.
delay_end (int) – maximum delay in search. default is 200.
- Kwargs:
plotting (bool): Plotting for user interpretation. defaut is False. normalize (bool): Normalizes the permutation entropy on scale from 0 to 1. defaut is False.
- Returns:
MsPE, the permutation entropy over multiple time scales.
- Return type:
(array)
Example:
import numpy as np
t = np.linspace(0,100,2000)
ts = np.sin(t) #generate a simple time series
from teaspoon.SP.information.entropy import MsPE
delays,H = MsPE(ts, n = 6, delay_end = 40, normalize = True)
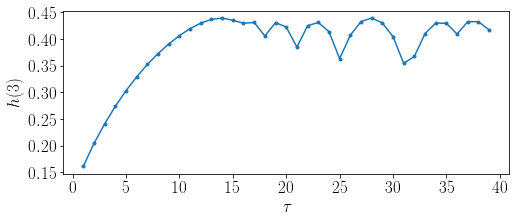
import matplotlib.pyplot as plt
plt.figure(2)
TextSize = 17
plt.figure(figsize=(8,3))
plt.plot(delays, H, marker = '.')
plt.xticks(size = TextSize)
plt.yticks(size = TextSize)
plt.ylabel(r'$h(3)$', size = TextSize)
plt.xlabel(r'$\tau$', size = TextSize)
plt.show()
Output of example:
2.4.3.1.3. Persistent Entropy
Persistent entropy uses the data types of lifetimes from a persistence diagram and calculates their respective probability as
where L is the set of lifetimes.
- teaspoon.SP.information.entropy.PersistentEntropy(lifetimes, normalize=False)[source]
This function takes a time series and calculates Permutation Entropy (PE).
- Parameters:
lifetimes (array) – Lifetimes from persistence diagram (1d).
- Kwargs:
normalize (bool): Normalizes the entropy on scale from 0 to 1. defaut is False.
- Returns:
PerEn, the persistence diagram entropy.
- Return type:
(float)
Example:
import numpy as np
#generate a simple time series with noise
t = np.linspace(0,20,200)
ts = np.sin(t) +np.random.normal(0,0.1,len(t))
from teaspoon.SP.tsa_tools import takens
#embed the time series into 2 dimension space using takens embedding
embedded_ts = takens(ts, n = 2, tau = 15)
from ripser import ripser
#calculate the rips filtration persistent homology
result = ripser(embedded_ts, maxdim=1)
diagram = result['dgms']
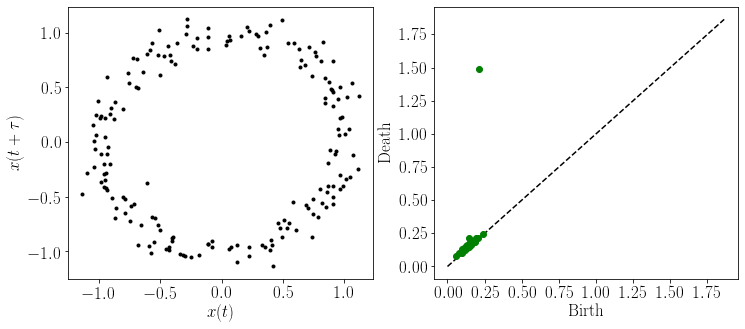
#--------------------Plot embedding and persistence diagram---------------
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
gs = gridspec.GridSpec(1, 2)
plt.figure(figsize = (12,5))
TextSize = 17
MS = 4
ax = plt.subplot(gs[0, 0])
plt.yticks( size = TextSize)
plt.xticks(size = TextSize)
plt.xlabel(r'$x(t)$', size = TextSize)
plt.ylabel(r'$x(t+\tau)$', size = TextSize)
plt.plot(embedded_ts.T[0], embedded_ts.T[1], 'k.')
ax = plt.subplot(gs[0, 1])
top = max(diagram[1].T[1])
plt.plot([0,top*1.25],[0,top*1.25],'k--')
plt.yticks( size = TextSize)
plt.xticks(size = TextSize)
plt.xlabel('Birth', size = TextSize)
plt.ylabel('Death', size = TextSize)
plt.plot(diagram[1].T[0],diagram[1].T[1] ,'go', markersize = MS+2)
plt.show()
#-------------------------------------------------------------------------
#get lifetimes (L) as difference between birth (B) and death (D) times
B, D = diagram[1].T[0], diagram[1].T[1]
L = D - B
from teaspoon.SP.information.entropy import PersistentEntropy
h = PersistentEntropy(lifetimes = L)
print('Persistent entropy: ', h)
Output of example:
Persistent entropy: 1.7693450019916783